Table of Contents
cs19f22as07: Protein Bioinformatics Lite
Goals
- Develop a class that represents some basic properties of the primary structure of proteins.
- Practice defining types that provide an idiomatic C++ interface.
Prerequisites
This assignment requires familiarity with the lecture materials presented in class through week 07.
Background
Proteins constitute a class of molecules that are the primary drivers of all biochemical activity in living organisms on Earth. Every viable cell in every Earth-based life form has DNA molecules inside of its nucleus, and through the process of protein biosynthesis, cells transcribe DNA into RNA, then translate that RNA into protein products.
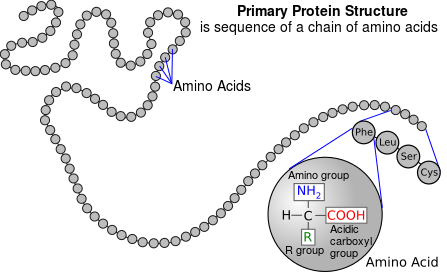
Protein biosynthesis involves assembling a chain of amino acids, each of which is coded for by a sequence of three RNA nucleotides. The resulting molecule is a form of polypeptide known as a protein. We refer to this sequence of amino acids as the primary structure of a protein. The secondary, ternary and quaternary structures refer to the complex shapes formed as hydrophobic interactions, intramolecular hydrogen bonds and van der Waals forces cause the primary structure of a protein to fold around itself and other proteins, giving the protein its essential biological functionality.
There are 20 proteogenic amino acids. We usually use single-letter abbreviations to refer to them (A for alanine, C for cysteine, etc.), constituting an alphabet for describing protein structure:
A C D E F G H I K L M N P Q R S T V W Y
File /srv/datasets/amino-monoisotopic-mass (also available via HTTP) contains a simple table of the monoisotopic masses (measured in daltons) of each of the 20 proteogenic amino acids, along with the letters that we usually use to refer to them. Here are the first five lines of the file:
A 71.03711 C 103.00919 D 115.02694 E 129.04259 F 147.06841
As in Rosalind problem PRTM, we can use this data to compute the weight/mass of a protein, given its primary structure. This information can be useful for the purposes of protein mass spectrometry, e.g. in the identification and categorization of proteins involved in diseases such as those produced by viruses like SARS-Cov-2, which is the foundation for developing mRNA vaccines such as those that were quickly developed in response to COVID-19.
Assignment
You shall define a class named Protein in the cs19 namespace, instances of which represent the primary structure of a protein using the amino-acid alphabet. For simplicity's sake, define the class entirely within file cs19_protein.h (i.e. no separation of interface from implementation this time).
cs19::Protein provides an idiomatic C++ interface, overriding various operators for the purposes of inspecting and modifying a cs19::Protein object, as well as computing its expected mass for the purposes of spectrometry.
Design your class to match this specificiation according to the description of each member function. You must implement at least stub versions of all of the specified constructors and member functions in order to receive a grade.
You will save yourself a substantial amount of work if start with a copy of the Dna class from week 07 and modify it to represent proteins instead of DNA sequences. Much of the code could conceivably be quite similar.
Testing
Here is a small amount of code that uses assertions to test the constructors and the mass() function, then reads strings from stdin, assumes those strings are protein descriptors, and prints a couple of facts about each protein. I've also created a sample executable named cs19_protein that is a working version of this program, if you'd like to consult it for testing purposes.
#include <cassert> #include <cmath> #include <initializer_list> #include <iostream> #include <stdexcept> #include <string> #include "cs19_protein.h" int main(int argc, char **argv) { // from Rosalind PRTM: https://rosalind.info/problems/prtm/ const std::initializer_list<char> prtm_prot{'S', 'K', 'A', 'D', 'Y', 'E', 'K'}; constexpr double prtm_mass = 821.392; assert(std::abs(cs19::Protein(prtm_prot).mass() - prtm_mass) < .001); assert(std::abs(cs19::Protein(std::string(prtm_prot)).mass() - prtm_mass) < .001); assert(std::abs(cs19::Protein(std::string(prtm_prot).c_str()).mass() - prtm_mass) < .001); assert(std::abs(cs19::Protein(prtm_prot.begin(), prtm_prot.end()).mass() - prtm_mass) < .001); cs19::Protein test; while (std::cin) { try { if (std::cin >> test) std::cout << test << ' ' << test.size() << ' ' << test.mass() << '\n'; } catch (std::domain_error &error) { std::cerr << error.what() << '\n'; } } }
e.g. try running the sample executable and giving it some text on stdin or the name of a file (such as /srv/datasets/ebola_orf_products which contains 220 protein descriptors from the segments of the Ebola virus reference genome that constitute open reading frames, or potential protein products of Ebola:
% cs19_protein <<<SKADYEK SKADYEK 7 821.392 % tail -5 /srv/datasets/ebola_orf_products | cs19_protein MGHGKLSLRNYQS 13 1471.74 MQDSEVKLIERLTGLLSLFPDGLYRFD 27 3136.63 MSLTQHNKLRTLYN 14 1699.88 MLYQYLARWNSALVDNTTS 19 2227.07 MTVKSEIPSLQYSRLDNNLRVNDN 24 2787.4
Submission
Submit cs19_protein.h via turnin.
 Feedback Robot
Feedback Robot
This project has a feedback robot that will run some tests on your submission and provide you with a feedback report via email within roughly one minute.
Please read the feedback carefully.
Due Date and Point Value
Due at 23:59:59 on the date listed on the syllabus.
Assignment 07 is worth 60 points.
Possible point values per category:
---------------------------------------
Correctly implemented member 60
functions, split roughly evenly
(constructors and to_string() must
work to receive much credit)
Possible deductions:
Repeated/no access to
amino-monoisotopic-mass 25%
Style and practices 10–20%
Possible extra credit:
Submission via Git 5%
---------------------------------------